![]()
Application of AI algorithms for off-target prediction
in CRISPR-Cas9 gene editing
“Report on the seminar given by Jeremy Charlier on February 22th 2022”
BIF7002
Written by:
Wasmi Algasim, Salwa Haidar, and Zeinab Sherkatghanad
UQAM - Winter 2022
table of contents
1. Introduction
2. CRISPR/Cas9 Genome editing
3. Application
of AI algorithms on target prediction
3.1 Sequence Encoding
3.2
Application of Machine
learning
3.3 Application of Deep
learning
4. Off-target
prediction with novel sgRNA-DNA sequence encoding in CRISPR-Cas9 gene editing
5. Conclusion
6. Reference
1.
Introduction
Discovery of genome editing
technology has revolutionized molecular biology and genetics.
Genome editing or gene
editing represents a set of approaches that permit the manipulation of the
genome of living organisms. Gene editing allows to make specific changes in DNA
sequences such as DNA insertion, deletion, modification of certain nucleotides
or replacement of a specific DNA fragment [1].
Gene editing techniques
rely on the use of particular enzymes called nucleases that can be directed to
target a specific DNA sequence and enable gene editing at the cut sites. DNA
nucleases induce double-strand breaks (DSB) at the DNA level that can be repaired
by one of the 2 major DNA repair mechanisms in the cell, homology directed
repair (HDR) and non-homologous end joining (NHEJ) repair, frequently
accompanied by indels (insertion/deletion) at the cut site [2].
Zinc-finger nucleases
(ZFNs) and transcription activator-like effector nucleases (TALENs) were the
two main strategies, based on protein-DNA interaction, used for gene editing
between 1985 and 2012 [3]
[4]
[5].
Genome editing
technologies have evolved rapidly in the past few years leading to the
discovery of a more powerful and efficient tool for DNA manipulation called
CRISPR/Cas9. CRISPR/Cas9 system has been launched in 2012 and it is based on
RNA-DNA complementarity, instead of protein-DNA interaction, for target DNA
sequence recognition. It represents the most popular gene editing technique
nowadays and it improved our understanding of several gene roles, biological functions
and genetical diseases [5]
[6].
Significant properties
of CRISPR-associated protein 9 (Cas9) such as flexibility, cost-efficiency,
simplicity and ability to remove more than one gene at a time have evoked great
enthusiasm on this technique. CRISPR/Cas9 as a superior gene editing
technology, has been rapidly developed in various research fields, ranging from
basic research on genetic therapies at the cellular level to applied biomedical
research [6]
[7]. Beside clinical
potential of CRISPR/Cas9 for treating human disease such as cancer, genetic
disorders and beyond [8] [9]
[10], it has efficient
achievements in genetic engineering for plants [11] and in animal disease
models [12]. A Schematic of
CRISPR/Cas9 genome editing system and its applications has been provided in Figure
1.
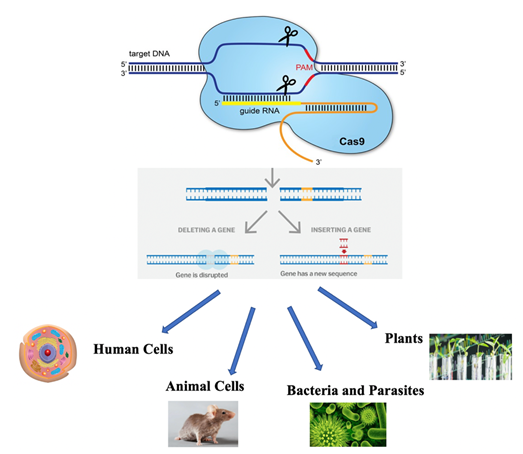
Figure 1. CRISPR/Cas9 genome
editing system and its application domains.
Adapted from [1].
2. CRISPR/Cas9 Genome editing
CRISPR-Cas9 is an adaptive
immune system used in some bacteria and archaea to defend themselves against
foreign DNA from invading viruses. CRISPR, stands for Clustered Interspaced
Short Palindromic Repeats, consists of a succession of
repeats separated by distinct sequences called “spacers” from viral genomes. Cas9 (CRISRP-associated protein 9) is an endonuclease
that cuts DNA within 20 nucleotides located immediately before a protospacer
adjacent motif (PAM) composed of 3 nucleotides (NGG) (Figure 1) [13]
[14].
CRISPR/Cas9 system has
two main components: a Cas9 protein responsible of cleaving the double strands
of DNA sequences and a single guide RNA (sgRNA), composed of 20 nucleotides,
directing the Cas9 to the target sequence in the genome and ensuring cutting
takes place in the right place [1]. The
Protospacer-Adjacent Motif (PAM) which is a 3 nucleotide motif located at the
end of the DNA target site, is required for Cas9 protein to cleave at a
specified site [15].
The CRISPR/Cas9 is quite accurate in gene editing. sgRNA can precisely edit the
target site (i.e. on-target editing), though it may bind and wrongly edit at
other additional sites leading to unintended off-target effects.
Therefore, the safety aspect
of CRISPR/Cas9 on humans is still an open issue and there are concerns about
the practical applications of this technique. It is thus highly desirable to
design data-driven models which;
(1) Influence on-target
efficiency
(2) Improve off-target
specificity
(3)
Simultaneously maximize on-target activity and minimize off-target effects
Over the last few years,
the main objective of data-driven models has been to predict of on-target and
off-target activity, and in all cases, investigating an informative encoding
technique and designing effective models for learning features is a major
challenge. These computational algorithms can improve our understanding of the
mechanisms of CRISPR/Cas9 system and provoke more enthusiasm on the clinical
application of this technique. A lot of tools and bioinformatic algorithms have
been developed to help in the design of the sgRNA and to check possible
off-target effects. A summary about some of these tools is presented in Table 1
[16].
Off-target effects are undesired and should be minimized.
|
Tool Name |
Search by Gene Name |
Alternate PAM Sequence |
Predicts Off-targets |
Ranks Output |
All in One Tool |
Link |
|
Cas9-Design |
× |
× |
✓ |
× |
✓ |
|
|
CCTop |
× |
✓ |
✓ |
× |
✓ |
|
|
CGAT |
✓ |
× |
✓ |
✓ |
✓ |
|
|
CHOPCHOP |
✓ |
✓ |
✓ |
✓ |
✓ |
|
|
COSMID |
× |
✓ |
✓ |
✓ |
✓ |
|
|
CRISPR design |
× |
✓ |
✓ |
✓ |
✓ |
|
|
CRISPRdirect |
✓ |
✓ |
✓ |
✓ |
✓ |
|
|
Crispr Finder |
× |
× |
✓ |
× |
× |
|
|
CRISPR
Multitargeter |
× |
✓ |
✓ |
× |
× |
|
|
Crispr-P |
✓ |
✓ |
✓ |
✓ |
✓ |
|
|
CRISPRseek |
× |
✓ |
✓ |
× |
✓ |
http://www.bioconductor.org/packages/release/bioc/html/CRISPRseek.html |
|
CROP-IT |
✓ |
✓ |
✓ |
✓ |
× |
http://cheetah.bioch.virginia.edu/AdliLab/CROP-IT/homepage.html |
|
E-crisp |
× |
✓ |
✓ |
× |
× |
|
|
flyCRISPR |
× |
✓ |
× |
× |
✓ |
|
|
GT-SCAN |
× |
✓ |
✓ |
× |
✓ |
|
|
sgRNAcas9 |
× |
✓ |
✓ |
× |
× |
|
|
SSFinder |
× |
× |
× |
× |
× |
Table 1: Tools for sgRNA design and off-target
effect prediction [16].
3.
Application
of AI algorithms on target prediction
In this section, we discuss the most important applications
of Deep Learning and Machine learning Methods for Genome Editing. On-target and
off-target predictions in the context of deep learning and machine learning can
generally be divided into two main challengeable parts. First, those that
obtain an encoding technique to convert sgRNA-DNA sequence pairs into an
appropriate input for machine and deep learning models without loss of
information. Second, those that design effective machine and deep learning
models to learn features from vector or matrix representations and provides
high accuracy predictions.
3.1 Sequence Encoding
Recently, several studies have been
conducted to use the sequence pair information effectively. In fact, one of the main
challenges in off-target prediction is converting sgRNA-DNA
sequence pairs into an appropriate input for deep learning models without
loss of information. In this case, several attractive encodings
techniques have been explored based on one-hot encoding and k-mer embedding.
In one-hot encoding,
each
sgRNA-DNA sequence pair encoded in a one-hot matrix with 4 rows correspond to
the size of nucleotide type, i.e. A, C, G and T, and L columns correspond to
the length of the sequence. Therefore, each base in in the sgRNA and target DNA
is encoded in the form of four one-hot vectors [1,0,0,0], [0,1,0,0], [0,0,1,0]
and [0,0,0,1]. There are many outstanding one-hot encodings technique to encode
sequence pairs. To cover this important encoding technique, we briefly
summarize the more recent studies and their used encoding schemes.
Lin et al [17]
introduced a novel one-hot sequence encoding technique to transfer each
sgRNA-DNA sequence pair with length of 23 (3-bp PAM adjacent to the 20 bases)
into a 4´23 matrix using the OR
operation on two one-hot vectors of base-pairing. Lin et al. [18]
proposed an encoding technique that each gRNA-target pair can be represented by
10´L binary matrix (L is the length of
sequence). They encode each gRNA target sequence pair with indels and
mismatches with five-bit one-hot encoding with channel-wise concatenation.
Stortz et al. [19],
investigated four feature encoding techniques called target-guide encoding,
target-mismatch encoding, target-mismatch-type encoding and target-OR-guide
encoding to implement physically informed features.
Chuai et al [20] presented an encoding
method to encode a DNA region which contains both nucleotide sequence and
epigenetic information. They considered DNA region as a one-row multi-channel
picture includes A-channel, C-channel, G-channel, and T-channel, and also each
epigenetic feature is considered as one channel. Zhang et al. [21] proposed a new encoding
scheme to encode sequence pairs with bulges without information loss in the
process of encoding. For different two bases in a base pair, an OR operation on
the two vectors is performed to represent the base pair. For same two bases in
a base pair, the encoding approach used OR operation and reverses it. Y. Zhang
et al. investigated a new encoding that each
sequence converted into 20´20 matrix.
They used concatenation on the one-hot features for on-target sequence, one-hot
features for putative off-target sequence, and the mismatch position. Since, the
mismatch positions include 12 different mutation type, mutation position and
type information can be represented by a 12´20
matrix.L.
Xue et al. [22]
used one-hot encoding on sgRNA on-target activity prediction.
Another important encoding technique is the k-mer
embedding, which is inspired by the word2vec technique [23]. In this encoding
approach, the input sequence is split into overlapped k-mers of length k using
a sliding window with stride s, then each k-mer is mapped into a d-dimensional
vector using word2vec method. Word2vec is an unsupervised learning algorithm
which maps k-mers from the vocabulary to vectors of real numbers in a
low-dimensional space.
Charlier
et al. [24]
represented a new one-hot encoding method based on One-hot encoding that maps
sgRNA-DNA sequence pair into 8´23 matrix according to a
bijective function. This encoding approach is based on the concatenation of the
sgRNA and the DNA nucleobase sequences and preserves information throughout the
encoding process (Figure 2).
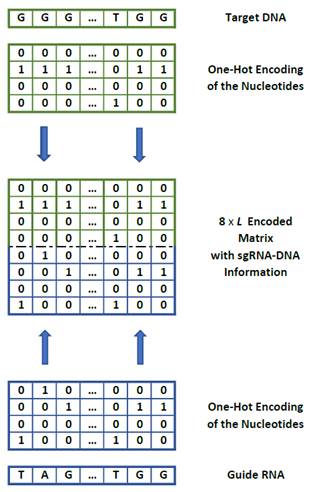
Figure
2. 8x23 encoding of guide-RNA and target DNA.
3.2 Application of Machine learning
In
CRISPR/Cas9, traditional machine learning algorithms has been studied
extensively for on-target and off-target prediction. In this section, we
provide a summary of the application of machine learning methods in genome
editing and their impacts on target predictions. Before the data driven model
was used as a common method in genome editing, some custom prediction methods
based on scoring functions such as MIT CRISPR Design Tool2, CCTop algorithm,
CRISPR Design, E-CRISP and CHOPCHOP were designed. These custom tools have
their own limitations and there are concerns that they may miss extra
information.
The
initial attempt to use a data driven model was provided in 2014 by Wang et al. [25] that
implement support-vector-machine (SVM) classifier. Then, and Doench et al. [26] proposed
SVM and Logistic regression.
3.3 Application of Deep
learning
Although CRISPR-Cas is a
powerful genome editing technology, there are concerns about the safety aspect of
its translational applications. Design of off-target prediction models to
evaluate the off-target situation of various gRNAs accurately has a great
impact on the selection of gRNAs with high specificity and targeting accuracy.
Lin et al. [17]
proposed a deep neural network for off-target predictions. They investigated
two deep convolutional neural network and deep feedforward neural network to
predict off-target mutations. Engineer SpCas9 with higher specificities plays
an important role to address the off-target problem. Wang et al [27], analyzed CNN and RNN
framework for gRNA activity prediction for Cpf1, WT-SpCas9 and SpCas9-HF1. The
RNN frame work have shown promising results compare to CNN and other
algorithms. They developed an online tool based on RNN called DeepHF to improve
on-target activity prediction. This tool, which combines RNN and important
biological feature (biofeature), is able to predict the activities of all gRNAs
with DNA sequence as input and select gRNAs that are suitable for gene knockout
with eSpCas9(1.1) and SpCas9-HF1 data.
Chuai et al. [20] implemented a novel
deep learning framework named DeepCRISPR to simultaneously predict sgRNA
on-target knockout efficacy and the off-target cleavage. They presented a deep
unsupervised representation learning approach to pre-train huge amounts of
unlabeled sgRNAs. In this framework,
first sgRNAs encoded with its sequence and epigenetic information and then a
deep coevolutionary denoising neural network (DCDNN) is presented to learn
meaningful representation of sgRNAs. Finally, the output is used as the input
of a coevolutionary neural network. Thus, the model not only trained the wights
of CNN but also fine-tuned the weights of DCDNN-based network with limited
labeled sgRNAs. Also, they extend this model for sgRNA off-target site
prediction by reusing the pre-trained DCDNN-based network.
Attention mechanism is
also effective to achieve a satisfactory performance for sgRNA off-target
specificity prediction and on-target efficiency prediction. Liu et al. [28] analyzed two
transformer-based deep neural network model called AttnToMismatch_CNN and
AttnToCrispr_CNN to consider cell-specific information of genes. These
attention-based models, AttnToMismatch_CNN and AttnToCrispr_CNN have
competitive performance for off-target sgRNA specificity prediction and
on-target efficiency prediction, respectively. Also, they implemented seqCrispr
model which compromise a LSTM component and CNN component in parallel for
on-target efficiency predictions. AttnToMismatch_CNN architecture contains four
components: an embedding layer (the base pairs at different positions are
encoded into distinct vector representations), a transformer layer, a
convolutional neural network layer and fully connected layer. AttnTo-Crispr_CNN
frameworks consist of four components the same as AttnToMismatch_CNN except
that AttnToCrispr_CNN has a linear regression layer for the final output.
4. Off-target
prediction with novel sgRNA-DNA sequence encoding in CRISPR-Cas9 gene editing
In this section, we
consider an interesting method for off-target prediction with novel sgRNA-DNA
sequence encoding in CRISPR-Cas9 gene editing. Charlier et al. [24]
propose the novel encoding method that has shown promising results for
off-target prediction. They considered two experiments, one on each data set.
The first is the CRISPOR dataset and the second is GUIDE-seq dataset. On both,
many types of deep learning models were tested, such as Convolutional Neural
Network (CNN), Recurrent Neural Network (RNN) and Feedforward Neural Network
(FNN) for example. Some of these tests were performed on different depth of
layers, and the best of each (i.e., ones that performs best according to their
F1-score) were picked and used to be compared against the best model of its
type from the other dataset. For example, FNN models tested in Charlier et al.
experiments were FNN 3 layers, FNN 5 layers and FNN 10 layers, meaning 3-5-10 connected
dense layers respectively. Using their CRISPOR dataset, FNN 5 layers for the
4x23 encoding was the best model in terms of performance. As for the 8x23
encoding, the FNN 10 layers was the best performing one. For the first test,
the model predicted 6 positive samples out of 43 positives, as for the latter
the top 20 predictions were true positive samples [24].
Figure 3 could be looked at to see which deep learning models were used.
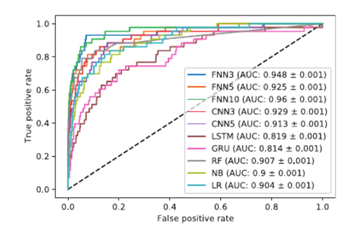
Figure 3. Roc curves (AUC) from
Charlier et al 2021 experiment using the 4x23 encoding on the CRISPOR
dataset [24].
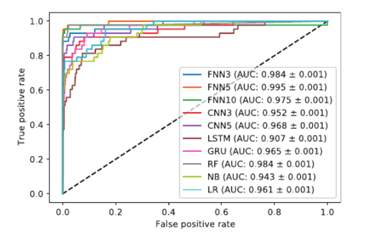
Figure 4. Roc curves (AUC) from
Charlier et al 2021 experiment using the 8x23 encoding on the CRISPOR
dataset [24]
To present the
performance of each model, the results in Charlier et al 2021 study were
presented are the form of Receiver Operating Characteristic [29] curves. It is clear
that this novel encoding method improves the accuracy of off-target prediction
on every model tested, with the highest ROC (AUC) score of 99.5% (Figure 4).
However, this method presented Charlier et al could be improved, for example,
if it took into consideration the possibility of insertions, deletions (indels)
in addition to mismatches to portray reality more reliably. It is reported by
them that indels could be considered by adding two additional rows to the
encoded matrix.
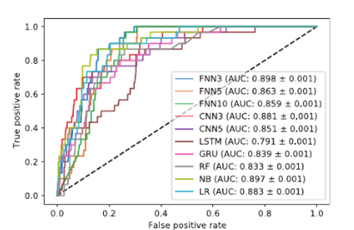
Figure 5. Roc curves (AUC) from
Charlier et al 2021 experiment using the 8x23 encoding on the GUIDE-seq
dataset [24].
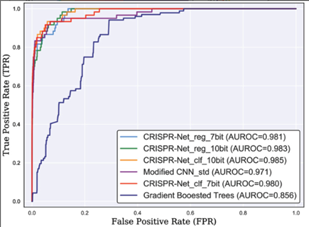
Figure 6. Roc curves (AUC) from
Lin et al 2020 experiment using the 7x24 encoding on the GUIDE-seq
dataset [18]
A study performed by Lin
et al 2020 use another method to consider indels. They have done that by
encoding the sgRNA-DNA sequences in a 7 x 24 matrix. However, the deep learning
model developed to score the mismatch and indels is a recurrent convolution
network which is a combination of inception-based convolutional neural network
and bidirectional LSTM (Lin 2020). When we compare the two highest scores from
Lin et al 2020 and Charlier et al 2021 on the same dataset (Figure 5
and Figure 6), which are FNN3 for the first and CRIPSR-Net-cif for the
second, we notice that the latter model outperforms the first one. This
analysis still must be taken with precaution, because it’s difficult to compare
two methods that share a few parameters.
5.
Conclusion
Deep Learning is a
powerful method for the target prediction and learning complex patterns at
multiple layers that has been used in many research works. Compared to
traditional machine learning methods, deep Learning have been applied to
computational biology to manage growing amounts of generated data. In the
experiment performed by Charlier et al 2021 [24],
the novel encoding method has shown promising results for off-target
prediction. They propose
a novel encoding method for off-target prediction in CRISPR/Cas 9 technique.
They consider CRISPOR and GUIDE-seq dataset for deep learning models such as
Convolutional Neural Network (CNN), Recurrent Neural Network (RNN) and
Feedforward Neural Network (FNN) for example.
Overall, the prediction
for off-target activities is still a successful step forward in research and
would enable better outcomes in the clinical usage. It would be interesting
explore a comparison of the model proposed by Lin et al. [18]
and the model proposed by Charlier et al. [24]
by adding 2 extra rows, as indicated earlier, to evaluate performance of
prediction including indels. Also, it is interesting to address the insertions
and deletions (indels) in the 8x23 encoding and use parameter optimization
method for better performance of deep learning algorithms.
References
1. Redman,
M., et al., What is CRISPR/Cas9? Arch
Dis Child Educ Pract Ed, 2016. 101(4):
p. 213-5.
2. Maeder, M.L. and C.A. Gersbach, Genome-editing Technologies for Gene and
Cell Therapy. Mol Ther, 2016. 24(3):
p. 430-46.
3. Chuang, C.K. and W.M. Lin, Points of View on the Tools for Genome/Gene
Editing. Int J Mol Sci, 2021. 22(18).
4. Gaj, T., C.A. Gersbach, and C.F.
Barbas, 3rd, ZFN, TALEN, and
CRISPR/Cas-based methods for genome engineering. Trends Biotechnol, 2013. 31(7): p. 397-405.
5. Adli, M., The CRISPR tool kit for genome editing and beyond. Nat Commun,
2018. 9(1): p. 1911.
6. Jinek, M., et al., A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial
immunity. Science, 2012. 337(6096):
p. 816-21.
7. Cho, S.W., et al., Targeted genome engineering in human cells with the Cas9 RNA-guided
endonuclease. Nat Biotechnol, 2013. 31(3):
p. 230-2.
8. Cong, L., et al., Multiplex genome engineering using CRISPR/Cas systems. Science,
2013. 339(6121): p. 819-23.
9. Mali, P., et al., CAS9 transcriptional activators for target specificity screening and
paired nickases for cooperative genome engineering. Nat Biotechnol, 2013. 31(9): p. 833-8.
10. Ma, H., et al., Correction of a pathogenic gene mutation in human embryos. Nature,
2017. 548(7668): p. 413-419.
11. Liu, H., et al., CRISPR-P 2.0: An Improved CRISPR-Cas9 Tool for Genome Editing in
Plants. Mol Plant, 2017. 10(3):
p. 530-532.
12. Zarei, A., et al., Creating cell and animal models of human disease by genome editing
using CRISPR/Cas9. J Gene Med, 2019. 21(4):
p. e3082.
13. Zhang, F., Y. Wen, and X. Guo, CRISPR/Cas9 for genome editing: progress,
implications and challenges. Hum Mol Genet, 2014. 23(R1): p. R40-6.
14. Ma, Y., L. Zhang, and X. Huang, Genome modification by CRISPR/Cas9. Febs
j, 2014. 281(23): p. 5186-93.
15. Shah, S.A., et al., Protospacer recognition motifs: mixed identities and functional
diversity. RNA Biol, 2013. 10(5):
p. 891-9.
16. Brazelton, V.A., Jr., et al., A quick guide to CRISPR sgRNA design tools.
GM Crops Food, 2015. 6(4): p.
266-76.
17. Lin, J. and K.C. Wong, Off-target predictions in CRISPR-Cas9 gene
editing using deep learning. Bioinformatics, 2018. 34(17): p. i656-i663.
18. Lin, J.C., et al., CRISPR-Net: A Recurrent Convolutional Network Quantifies CRISPR Off-Target
Activities with Mismatches and Indels. Advanced Science, 2020. 7(13).
19. Florian Störtz, J.M., Peter Minary, piCRISPR: Physically Informed Features
Improve Deep Learning Models for CRISPR/Cas9 Off-Target Cleavage Prediction.
20. Chuai, G., et al., DeepCRISPR: optimized CRISPR guide RNA design by deep learning.
Genome Biol, 2018. 19(1): p. 80.
21. Zhang, Z.R. and Z.R. Jiang, Effective use of sequence information to
predict CRISPR-Cas9 off-target. Comput Struct Biotechnol J, 2022. 20: p. 650-661.
22. Xue, L., et al., Prediction of CRISPR sgRNA Activity Using a Deep Convolutional Neural
Network. J Chem Inf Model, 2019. 59(1):
p. 615-624.
23. Tomas Mikolov, I.S., Kai Chen, Greg
Corrado, Jeffrey Dean, Distributed
Representations of Words and Phrases and their Compositionality, 2013.
24. Charlier, J., R. Nadon, and V.
Makarenkov, Accurate deep learning
off-target prediction with novel sgRNA-DNA sequence encoding in CRISPR-Cas9
gene editing. Bioinformatics, 2021.
25. Wang, T., et al., Genetic screens in human cells using the CRISPR-Cas9 system.
Science, 2014. 343(6166): p. 80-4.
26. Doench, J.G., et al., Rational design of highly active sgRNAs for
CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol, 2014. 32(12): p. 1262-7.
27. Wang, D., et al., Optimized CRISPR guide RNA design for two high-fidelity Cas9 variants
by deep learning. Nat Commun, 2019. 10(1):
p. 4284.
28. Liu, Q., D. He, and L. Xie, Prediction of off-target specificity and
cell-specific fitness of CRISPR-Cas System using attention boosted deep
learning and network-based gene feature. PLoS Comput Biol, 2019. 15(10): p. e1007480.
29. Elzamly, S., et al., Epithelial-Mesenchymal Transition Markers in Breast Cancer and
Pathological Responseafter Neoadjuvant Chemotherapy. Breast Cancer (Auckl),
2018. 12: p. 1178223418788074.